Newsletter
The significance of ATP concentration in cell-free and cell-based assays

Carna Biosciences Newsletter Vol. 4
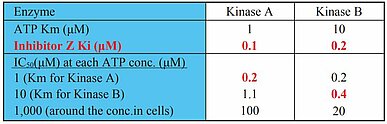
IC50 value - a direct measure of affinity between the inhibitor and the investigated kinase
In consideration of this difference, let’ s review the rationale of using ATP concentrations around the Km in cell-free assays. Km values approximate the affinity between ATP and the kinase, while Ki values describe the affinity between inhibitor and kinase. For ATP-competitive inhibitors, the dependencies between the half maximal inhibitory concentration (IC50) and ATP concentrations are described by the Cheng-Prusoff equation (IC50 = Ki + Ki/Km×[ATP]). An inhibitor has different Ki values for every kinase, and every kinase has a different Km for ATP. By representing the ATP concentration with its Km value, the IC50 reflects 2×Ki value. Thus the IC50 value becomes a direct measure of affinity between the inhibitor and the investigated kinase (1). Table 1 is an example where the IC50 of Inhibitor Z is calculated using the Cheng-Prusoff equation. It hypothesizes that the ATP Km values for Kinase A and Kinase B are 1μM and 10μM, respectively, and the Ki values of Inhibitor Z for Kinase A and Kinase B are 0.1μM and 0.2μM, respectively. The IC50 for Kinase A and Kinase B are calculated as 0.2μM and 0.4μM respectively when ATP concentration was assumed to be each Km value. This result reflects IC50 = 2×Ki value, thus establishing that the binding affinity of the inhibitor against various kinases can be ranked on the basis of IC50.
Comparison of cell-based vs cell-free assay
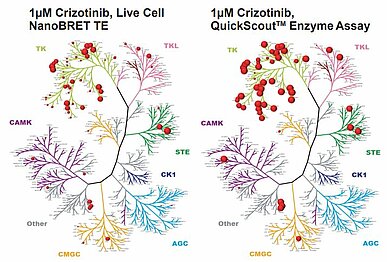
While this information is useful, prior to advancing a drug candidate, its cellular effects must be understood. Cellular ATP concentration is generally thought to be a few mM, and consistently higher than the ATP Km of most kinases. Table 1 shows that the IC50 values for Kinase A and Kinase B are calculated to be 100μM and 20μM at 1mM ATP, based on the Ki values. This illustrates that at high ATP concentrations, the IC50 discrepancies are greater for kinases with lower Km values compared to those with higher Km values. Robers M reported that the target occupancy of crizotinib at 1μM decreased drastically in a cell-based assay compared to the inhibition in a cell-free assay (2) (Fig.1).
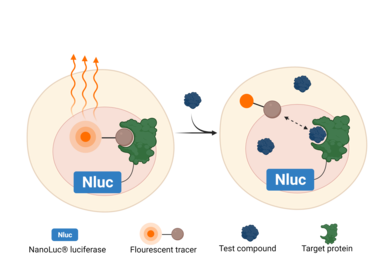
ATP concentrations in cell-free assays can be increased to more closely approximate cellular levels, however ATP concentration is not the only variable between these 2 assay systems. A multitude of factors affect compound binding affinity in cells. Given that cell membranes act as a barrier to binding, it is critical to confirm whether the inhibitor chosen by a cell-free assay maintains its inhibitory activity in a cell-based assay as well. To address this need, Carna Biosciences offers a cell-based assay service known as the NanoBRETTM Target Engagement (TE) Intracellular Kinase Assay System (Promega), in addition to cell-free recombinant kinase products and services (Profiling). In our cell-free assay service, we offer not only testing at ATP Km values but also at 1mM ATP, while our cell-based assay services can examine compound affinity to the target kinase in a cell. The target kinase fused with NanoLuc® luciferase can be expressed in cultured cells and the binding of test compounds to the kinase can be detected by BRET (Bioluminescence Resonance Energy Transfer) (Fig.2). Our cell-based assay makes it possible to evaluate the binding affinity of a compound to the target kinase inside intact cells, as it is performed without disrupting cell membranes. Additionally, it enables the determination of residence time, which is especially important as a new pharmacodynamic parameter. Residence time data is emerging as a highly effective tool to differentiate new drug candidates from known drugs and other compounds which may have the same IC50.
References:
- Protein Kinases as Drug Targets. 2011, ISBN: 978-3-527-317 90-5, B. Klebl.
- SLAS2020, Broad Kinome Selectivity and Residence Time Analysis in Live Cells with NanoBRET, Robers M.
- Cell Chem Biol 2018; 25(2): 206-214.e11. Vasta JD.
- PLoS One. 2014; 9(3): e92146. Uitdehaag JC.
Further news
-
Newsletter
Cancer treatments using inhibitors of CDK, a cell cycle regulator
Read moreCarna Newsletter Vol.16
-
Newsletter
ALK drug resistant mutations: Challenges for the treatment of lung cancer
Read moreCarna Newsletter Vol.15
-
Technical Note
DGKα and DGKζ are key targets for cancer immunotherapy
Read moreCarna Newsletter Vol.14
